Biological Optical Microscopy Platform
The Biological Optical Microscopy Platform (BOMP) makes state-of-the art fluorescence microscopy equipment available to the students and staff of the University of Melbourne, as well as the wider community.
-
Applications
Optical microscopy and Image analysis applications supported by BOMP
View -
Instruments
Complete list of BOMP instruments (link to specification sheets, booking calendar, instruction manual, responsible BOMP staff) and charging rates.
View -
Online Resources
Link to our Seminars & Workshops materials and recordings
View
Other platforms
The platform can be used in conjunction with:
- Australian Phenomics Network Histopathology and Slide Scanning Services
- Electron Microscopy (Ian Holmes Imaging Center) @ Bio21
- Electron Microscopy @ Bioscience
- Materials Characterisation and Fabrication Platform
- Melbourne Cytometry Platform
- Melbourne Histology Platform
- Melbourne TrACEES Platform
For more information about the wider platform community, please visit Research Website, which is available to all visitors.
Details about the University’s research infrastructure related resources and services can be found on Research Gateway, which is available to the University of Melbourne staff and Graduate Researchers.
Please read our Terms and Condition before accessing our Platform resources.
-
Training Registration - Microsope
In order to gain access to BOMP equipment, new users will need to go through a registration & training process.
View -
Training Registration - Image analysis/software
You can request training/consultation on image analysis and analysis software.
View -
Usage Charges
Find details of equipment usage charges
View -
Location
List of maps of our facilities
View
Access to the booking calendar will be given after the registration and training is completed.
Every instrument booking is done via iLab (BOMP landing page).
Registering to iLab
| Who | To do | Instruction |
|---|---|---|
Researcher (trainee) |
| UoM user (pdf/video) External user (pdf) |
Supervisor - with UoM finance account* holder |
| Supervisor with UoM finance account (pdf/video) |
Supervisor - Anyone without UoM Themis account |
| Supervisor without UoM finance account (pdf) |
*UoM Themis finance account is the internal finance account code within the UoM. Your supervisor should have it if their research fund is administered by the University of Melbourne. The Themis code should take the below pattern.
xx-xxxx-xx-xxxx-xxxxxxx-xxx-xx-xx (e.g. 01-5210-10-7236-063597-GEN-12-01)
A full UoM Themis code is required for accessing internal rate in iLab.
Additional resources
Additional resources can be found in Research Platform Booking including short tutorial video as below,
Calendar link
| Bio21 Institute | Bioscience |
| Zeiss Elyra LSM880, Leica SP8 | Nikon A1R, Nikon C2, Nikon Spinning Disk |
| Medical Building | Peter Doherty Institute |
| Zeiss LSM900, Zeiss LSM880, Zeiss LSM800, Operetta, UltramicroscopeII, Zeiss Axioscan7 | Zeiss LSM980, Zeiss LSM780, Thorlab Bergamo II , Olympus FVMPERS |
| Physics | Medicine (EHP) |
| Olympus FV3000 | Leica Stellaris 5 |
| Image analysis computers | Image analysis software |
| Diamond, Gold, Green, Cyan, Cherry | Imaris, Volocity, Huygens, Imaris stitcher |
BOMP Contact details
Phone: (03) 9035 5811
- Dr Paul McMillan

Manager, BOMPDr McMillan completed his PhD at the University of Glasgow in 2007. His research focussed on the molecular & cellular biology of the malaria parasite. He has wide expertise in the application of fluorescence and super-resolution microscopy techniques in medical research
- Dr Ellie Hyun-Jung Cho

Applications Specialist, BOMPDr Cho completed her PhD in the Dept. of Anatomy and Neuroscience at The University of Melbourne in 2009. Her research focus was on the ion channels in sensory pathways using fluorescence imaging, immunohistochemistry, and electrophysiology of neurons. She joined platform in 2014 and provides training and advice on image analysis and high-content imaging
- Dr Gabriela Segal

Applications Specialist, BOMPDr Segal has a PhD in Tumour Immunology from the Dept. of Microbiology and Immunology at the University of Melbourne. She has years of experience in sample preparation for confocal and multiphoton microscopy. In her current role as an academic specialist she supports confocal, tissue clearing and Lightsheet applications.
- Kalyan Shobhana

Support Officer, BOMPKalyan Shobhana completed her Masters in Biotechnology at The University of Melbourne. She has a background in biomedical science and optical microscopy, with a focus on confocal microscopy. In her current role as a Platform Support Officer at the Biological Optical Microscopy Platform, she is responsible for training and research support on the confocal microscopes.
- Jinny Chang

Support Officer, BOMPJinny Chang obtained her Bachelor of Biomedical Science (Honours) from Monash University. With a keen interest in developmental biology, she specialises in confocal and widefield microscopy techniques. In her current role as a Platform Support Officer at the Biological Optical Microscopy Platform, she is responsible for training and research support on the confocal microscopes.
- Dr Sangwon Yoo

Support Officer, BOMPSangWon Yoo completed her PhD in the Department of Anatomy and Physiology at the University of Melbourne in 2023. Her research focused on investigating the role of neuronal BDNF-TrkB signalling in regeneration of myelin in a murine model of Multiple Sclerosis. She has experience in confocal microscopy, spectral unmixing, and airyscan imaging. In her current role as a support officer at BOMP, she manages researcher training and provides training on confocal microscopes.
The Biological Optical Microscopy Platform is here to assist your research. If you have any questions then we urge you contact us bomp-enquiries@unimelb.edu.au.
In the meantime, here are answers to some of our commonly received questions over six categories: Access, Training, Registration, Building Access, Service, Data Management.
Access FAQs
-
How can I access BOMP service?
In order to gain independent access to BOMP equipment, new users will need to go through a registration and training process. See processes explained for
You can also access our other services. See Service FAQs for detail.
-
I have used the same instrument elsewhere. Do I still need to be trained?
Yes, any researchers new to our Platform SHOULD go through a registration and training process.
-
I'm trained but haven't used the instrument for a while, what should I do?
If you haven't used our system for longer than 6 months, you recommend have your first coming back session as a ‘refresher’ session. This is not a formal training but a semi-supervised imaging session with your own sample. This refresher session will be charged at the user operated (standard) rate.
Having refresher means BOMP staff will be around during your session to answer your question and making sure you can get back on to the system smoothly. This includes checking your access to facility/booking calendar, updating you on any changes of local procedure, going though instrument operation if necessary.
You can request a refresher session using our general instrument training request form. Please note that this requires iLab account (if you don't have iLab account yet, please complete step 1 on the registration process)
-
I am not interested in getting trained on the microscopes but would prefer if someone runs the samples for me is this possible?
We can perform a limited number of imaging sessions for researchers (2-3 sessions by prior arrangement). These sessions aren’t charged at a higher rate, but platform staff should be included in any outputs resulting from this work. Please see our Authorship Policy.
Training FAQs
-
How can I arrange a training session?
You can request training via iLab. See processes explained for
-
Can my supervisor train me on the microscope?
No. Only BOMP staff should perform trainings on the instruments. Once you have independent access to the system, you supervisor or colleagues can assist with how to acquire the data you require for your project.
-
When should I organise training?
You should organise training close to the time (within 1-2 weeks) that you are planning on imaging your own samples. This will give sufficient time to register and arrange the two training sessions required to get access to the microscopes. Be aware that on some busier microscopes, we may need to book session further in advance.
-
How many people can attend a group training?
First session : Pre-COVID19 restriction, we could train up to 3 people in the first session. This is to enable all the trainees to be able to see the computer screen and have a chance to get hands-on with the system. Under the COVID19 restriction, we can train single user at a time (no group training).
Second session: The second training must be one-on-one at all time as this involves optimising experimental parameter for user's own sample.
-
What is the charging for the training?
Approx $200-250 depending on your affiliation. The training is charged at a higher rate for the duration of your first training session (normally 2 hours). Please check our rate table for more detail as each system and affiliation has a different charge. In group trainings, these charges are shared equally across the trainees.
-
When will I get independent access to the instrument?
This usually happens after the second training session, once we have deemed you competent to use the system properly and independently. Additional training may be needed for some systems/researchers. Your trainer should confirm this when you are being trained.
Registration FAQs
-
What is a Themis code and where can I find it?
UoM Themis account is the internal finance account code within the UoM. Your supervisor should have it if their research fund is administered by the University of Melbourne. The Themis code should take the below pattern.
xx-xxxx-xx-xxxx-xxxxxxx-xxx-xx-xx (e.g. 01-5210-10-7236-063597-GEN-12-01)
A full UoM Themis code is required for accessing internal rate in iLab.
If your supervisor has valid Themis code, they should register iLab as UoM group leader using UoM email address. Then request University Service to add Themis account to their account. Once the Themis account is added in iLab, they should assign appropriate Themis code to their lab members. (see full instruction here).
If your supervisor cannot provide a Themis code, then they will be charged as an external researcher. External users are required to provide an invoice contact (name, email & phone) and a mailing address. Researchers from Monash University/Hudson/CSL require a Purchase order (PO) number before access can be provided.
-
What is OGTR? Does my sample require a PC2-certified lab?
Work on certain projects which deal with viable genetically modified organisms (GMOs) or human pathogens (RG2) are required to be performed in Physical Containment 2 (PC2) laboratories.
GMO: Projects involving GMOs are regulated by the Office of the Gene Technology Regulator (OGTR). Your lab will have an OGTR agreement that covers your project, which states what types of dealings your work falls under (your supervisor or lab manager should have this). Commonly, work that falls under a NLRD (notifiable low risk dealing) is required to be performed in a PC2 lab. More information on the types of GMO dealings can be found on this webpage.
Human Pathogens: Work on organisms that fall under the Risk Group 2 category (e.g. pathogenic E. coli, Campylobacter spp, Plasmodium spp, prions, HIV-infected blood) are required to be performed in a PC2 laboratory. More info on biosafety requirements can be found on this webpage.
If your GMO or pathogenic sample is fixed or falls under an exempt dealing then there is no requirement for additional OGTR paperwork.
-
My project involves a viable GMO or pathogenic sample. What do I need to do?
Researchers planning to bring viable GMO or pathogenic samples (Risk Group 2 which fall under a NLRD agreement) SHOULD do the following:
- Add the relevant BOMP facility to the NLRD agreement by emailing biorisk-management@unimelb.edu.au. Upon receiving confirmation, provide email evidence to BOMP.
- Provide the date that they have attended an "Complying with Gene Technology Requirements" session run by OREI.
- Read and complete SOP/RA#8 covering GMO/RG2 sample transport and decontamination of spills and provide to BOMP.
-
I am unable to access my iLab account using the UoM single sign on credentials. Who do I contact for support?
When you receive any error when logging into iLab using the UoM single sign on credentials, contact the Service Centre using the +61 3 834 40888/40999 or visit the University Services Portal. To help resolve the issue faster, please provide a screenshot of the error message.
-
I recently requested access to join a lab group on iLab. Who should I contact if my request was still pending?
All requests to join a lab group are sent to the Group leader or the Lab Delegate of the Lab Group. To assist with the request please contact your Supervisor or Lab Delegate to approve your request. See our booking page for instruction.
-
As a supervisor, what should I do if my staff/student requires iLab account setup?
If you hold UoM Themis account,
- You should be registered as a group leader in iLab (using your UoM email).
- Add your lab member to your lab group.
- Assign correct Themis account
If you do not hold UoM Themis account,
- You should be registered as a group leader in iLab (using non-UoM institutional email).
- Make sure to provide complete invoice detail during registration.
- Add your lab member to your lab group.
See our booking page for detail instruction.
Building Access FAQs
-
Can I get inducted into the BOMP facility without building induction?
No, all researchers performing independent research are required to undergo a building induction. Each building will have specific procedures which is explained in our iLab training request form. Please note that access to the facility will be given AFTER instrument training is completed.
-
How do I get access for afterhours usage in the Medical Building?
For certain instruments located at the Medical building, researchers are required to gain 20 hours experience on the instruments before they can apply for after-hours access (check local instruction of specific instrument).
Contact bomp-enquiries@unimelb.edu.au to arrange after-hours induction.
- Afterhours access to the instrument booking calendar: will be given after the induction by BOMP staff.
- Afterhours access to the Medical building: If you don't have it already on your student/staff card, you should complete Working in Isolation form and return to Medical Building reception.
- Afterhours access to the BOMP facility: If you don't have it already on your student/staff card, please bring your Medical Building Access Request form for BOMP staff to sign. You can retrieve your own form from the Medical Building reception and return afterwards.
-
How do I get access for afterhours usage in Bio21 institute?
Only researchers whose primary work is located within Bio21 will be able to be provided with after-hours access to the instruments. They will need to fill in the relevant working in isolation form and get it signed by their supervisor.
Service FAQs
-
Can BOMP help me with my sample preparation?
Yes, we can advise on how best to prepare your samples to get the best results. You can discuss with us during your training or contact us (bomp-enquiries@unimelb.edu.au) anytime.
-
Can BOMP help me on image acquisition (fee-for-service)?
We can perform a limited number of imaging sessions for researchers (2-3 sessions by prior arrangement). These sessions aren’t charged at a higher rate, but platform staff should be included in any outputs resulting from this work. Please see our Authorship Policy.
-
Can BOMP help me on replicating a method from a paper?
Yes, we can advise which systems are available to do this and advise how best to go about the experiment. We are also available to work collaboratively to help you establish methods that use some of our high-end microscopes (e.g. Lightsheet, High-content Imaging, FLIM, Image analysis).
-
Can BOMP help me on my image analysis?
Yes, we are available to assist with image analysis. We suggest that you have this meeting ahead of acquiring your data. We can also provide advice on how best to analyse your data, provide training on the relevant software and can even set up custom analysis scripts (via collaboration). Please see our collaboration policy.
-
I am writing a grant but unsure of the technical details of a new technique. Can BOMP help me with writing my grant?
Yes, we often assist with technical suggestions or proof-reading of researchers grants to ensure that they are correctly written. We can provide quotations or letters of support upon request. We can also assist with getting provisional data for inclusion in grants. Please see our collaboration policy.
-
Can BOMP help me with writing my manuscript?
Yes, we can assist with writing technical details in your methods & materials sections or how to present/interpret your data. Please see our collaboration policy.
Data Management
-
How do I access my data from BOMP instrument?
You can transfer your data from the acquisition computer to your final storage via Mediaflux Data Mover. Instruction is available here.
Workflow
- Once image acquisition is finished on the microscope, upload your data using Mediaflux DataMover installed on the acquisition PC.
- File upload will be initiated and run in the background. (i.e. you don’t have to wait. If the computer is turned off during upload, it will resume next time it is turned back on)
- When the upload is completed, you will receive an email notification and the download link.
- You can download the data to your final storage. Please note that the link will expire & Data will be deleted after 3 months.
-
Can I use USB drive/External hard drive to copy my data from microscope computer?
No, we are strictly preventing you from connecting any external hard drive/USB drive to the microscope acquisition computer to protect it from malware/virus. Data should be retrieved only via network (e.g. Mediaflux Data Mover).
-
How often do I need to back up my data from BOMP equipment?
You should back up your data immediately at the end of your session using Data Mover.
Acquisition computer is neither for storage nor for back up and the files accumulated in local drive will be deleted regularly by BOMP staff or accidently by other users. For regular clean up, notification email will be sent around around 2 weeks before, however, shorter time may be given if urgent. In addition, local files will NOT be backed up by BOMP and the researcher is responsible for archiving their own data. See our Data Management Policy for detail.
BOMP image competition 2022
Winner of 'Top 3 images'
-
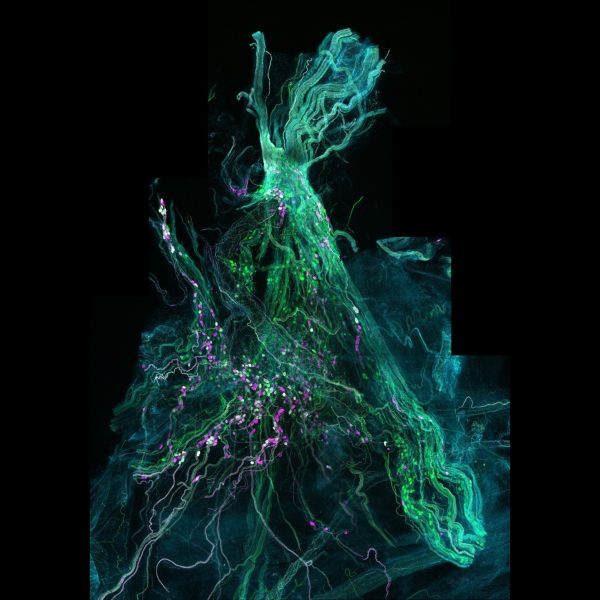
Mighty pelvic ganglion by John-Paul Fuller-Jackson, LSM900
Title: Mighty pelvic ganglion by John-Paul Fuller-Jackson Description: The major pelvic ganglion of the rat showing the various populations of neurons that reside within it. The nerves coming out of this ganglion are critical for pelvic organ function. Instrument:LSM900 -
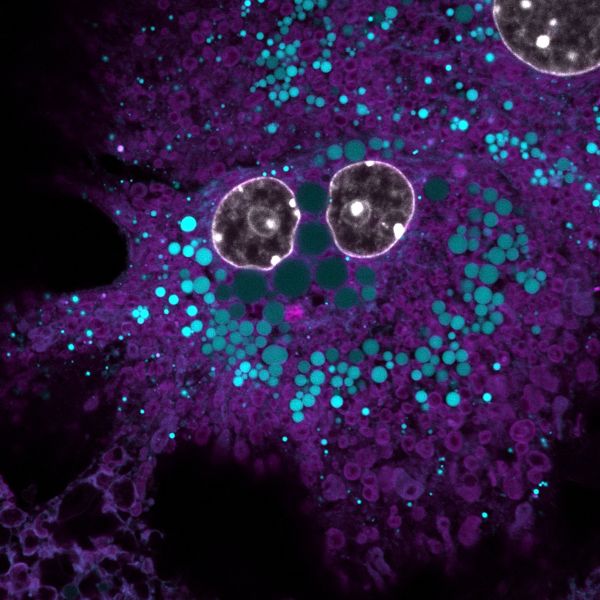
Smilin' cell-fie by Stacey Keenan, LSM900
Title: Smilin' cell-fie by Stacey Keenan, Description: Primary mouse hepatocyte stained with DAPI, BODIPY and MitoTracker to visualise nucleus, lipid droplets and mitochondria Instrument: LSM900 -
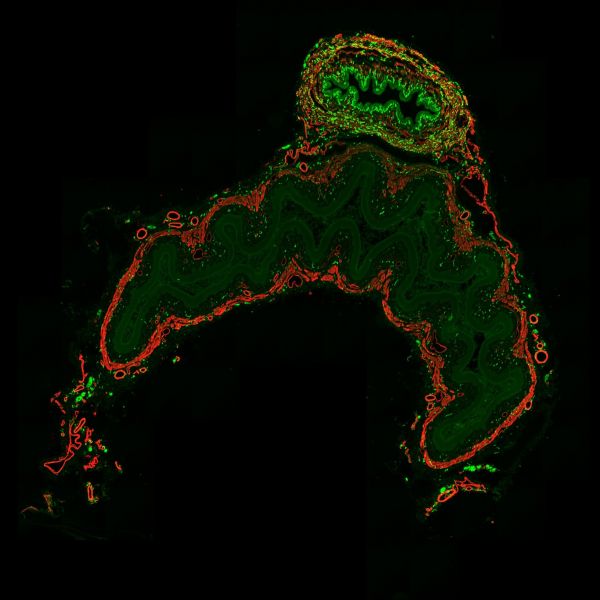
Unconventional Christmas Tree by Jonathan Chan, Axioscan7
Title: Unconventional Christmas Tree by Jonathan Chan Description: Christmas tree with red and green lights, plus a round shining star at the top. Transverse section of rat lower abdomen stained for CGRP axons (green), smooth muscle (red). Instrument: Axioscan 7
Note: Image size was reduced for some images.
Submitted entries
-
FLIM HeLa cell by Soheila Sabouri, FV3000
Title: FLIM HeLa cell by Soheila Sabouri Description: It is a FLIM result of HeLa cells stained by new generation of fluorescent probes (AIE: Aggregation-Induced Emission). Lifetme of the dye in every part of cell is different which shows the combination of this AIE dye and the imaging technique is successful to map and measure intracellular microenvironment accurately. Instrument:FV3000 -
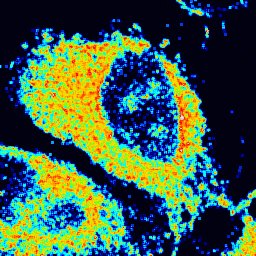
Anisotropy FAIM by Soheila Sabouri, FV3000
Title: Anisotropy FAIM by Soheila Sabouri Description: Fluorescence Anisotropy Imaging microscopy of HeLa cells stained by new generation of fluorophores (AIE: aggregation-induced emission). Instrument:FV3000 -
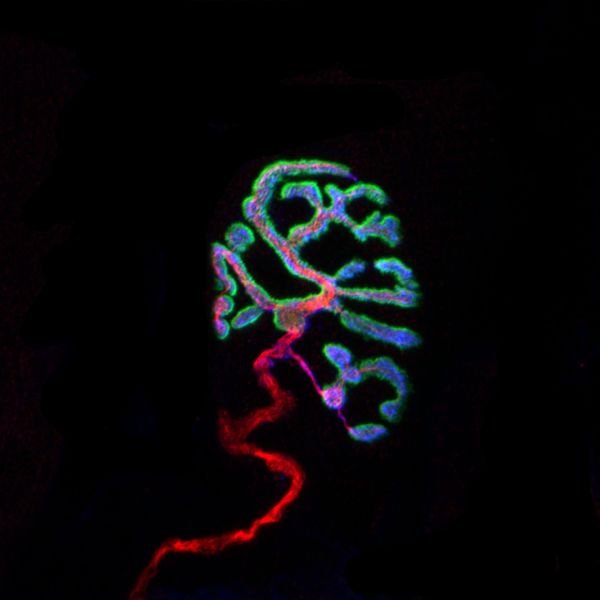
Neuromuscular Junction by Chris Karagiannis, LSM900 Airyscan
Title: Neuromuscular Junction by Chris Karagiannis Description: This is a composite image of a neuromuscular junction. Every muscle fiber is innervated by a single neuron and at the intersection of this innervation forms the displayed junction. Red: microtubules following the axonal body. Blue: synaptic vesicles localized to the nerve terminal. Green: acetylcholine receptors at the muscle endplate. Instrument:LSM900 Airyscan -
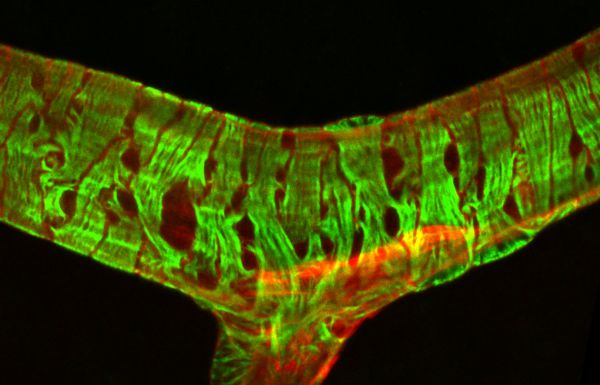
Bandaged vessel by Pialuisa Quiriconi, LSM800
Title: Bandaged vessel by Pialuisa Quiriconi, LSM800 Description: The retina is the light-sensitive tissue sitting at the back of the eye and is responsible for vision. This image depicts a large blood vessel (red) in the retina and the contractile cells (green) wrapping around it like bandage, that control vessel tone to regulate blood flow. -
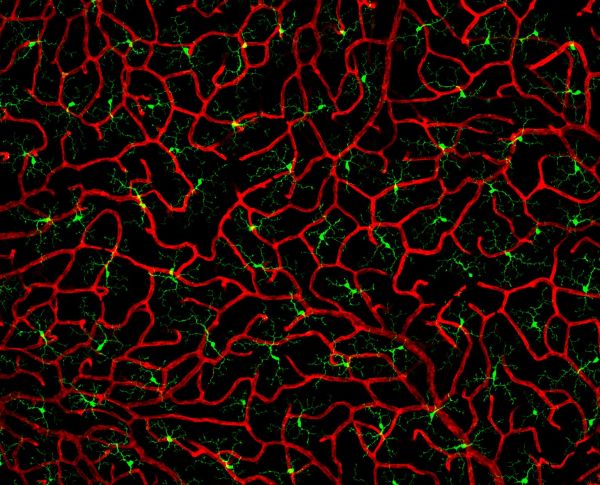
Coral in the eye by Pialuisa Quiriconi, LSM800
Title: Coral in the eye by Pialuisa Quiriconi, LSM800 Description: The retina is the light-sensitive tissue sitting at the back of the eye and is responsible for vision. This image depicts the finest blood vessels (red) in the retina called capillaries. Many cells reside amongst this dense network of vessels that resembles coral, including immune cells called microglia (green). -
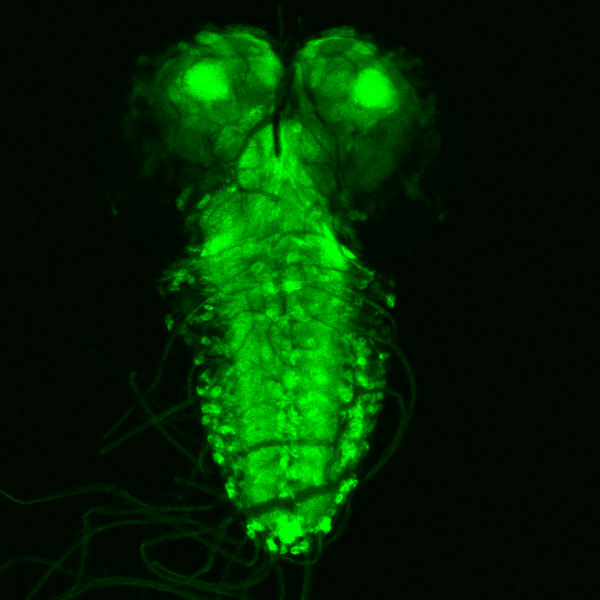
Gaze of the abyss by Wei Chen, Leica SP8
Title: Gaze of the abyss by Wei Chen, Leica SP8 Description: The image shows the expression of an important neuronal receptor from Drosophila larval brain, that is targeted by the most widely used insecticide imidalorpid, which provide better understanding towards the insecticide's mode of action and help improve the design of insecticides of the future. -
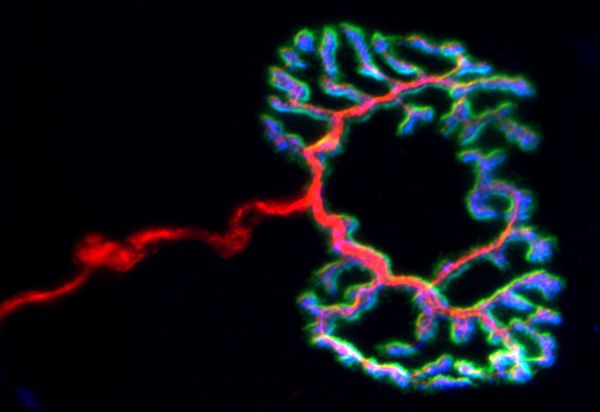
Neuromuscular Junction by Chris Karagiannis, LMS900
Title: Neuromuscular Junction by Chris Karagiannis, LMS900 Description: The intersection between a neuron and the muscle fiber being innervated forms the imaged neuromuscular junction. -
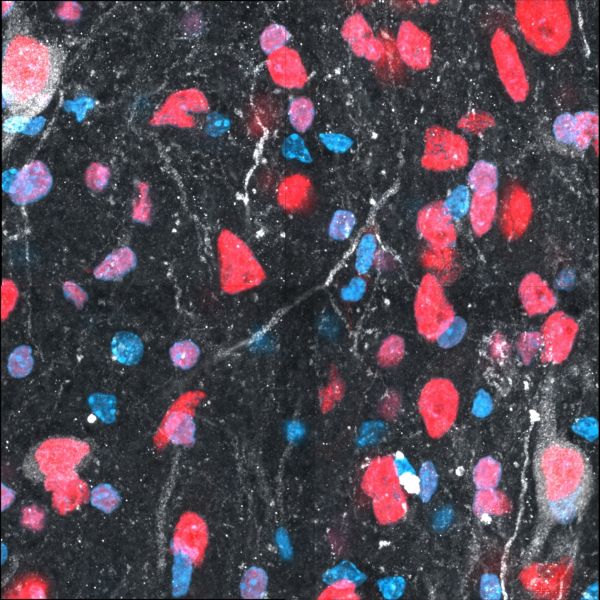
The Ghost by Chau Le Bao Tran, LSM900
Title: The Ghost by Chau Le Bao Tran, LSM900 Description: Transplanted human cells (cell body in white and human nucleus in red) in mouse brain (DAPI in blue). Important for drug screening on human tissue in living host -
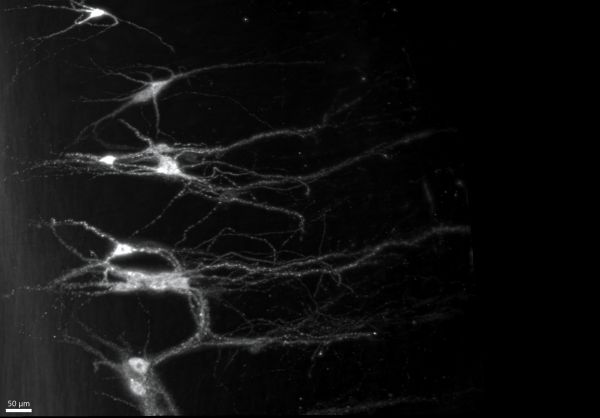
A few motoneurons by John-Paul Fuller-Jackson, Ultramicroscope II
Title: A few motoneurons by John-Paul Fuller-Jackson, Ultramicroscope II Description: Skeletal muscle is controlled by motoneurons in the spinal cord, which integrate signals from the environment and the brain via the spread of fibers seen in the image known as dendrites. -
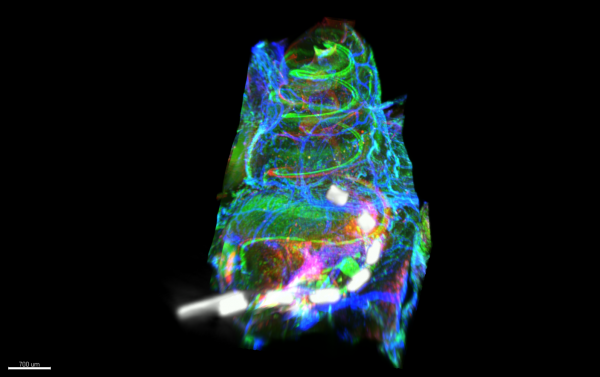
Cochlear Implant in situ by Kate Brody, Ultramicroscope II
Title: Cochlear Implant in situ by Kate Brody, Ultramicroscope II Description: Light sheet microscopy image showing a cochlea which is implanted with a cochlear implant. Hair cells and macrophages are labelled in the green, neurons and extracellular matrix in the red and arterioles and myo-fibroblasts in the blue. Co-registered with the image is a micro-CT of the cochlear implant in situ -
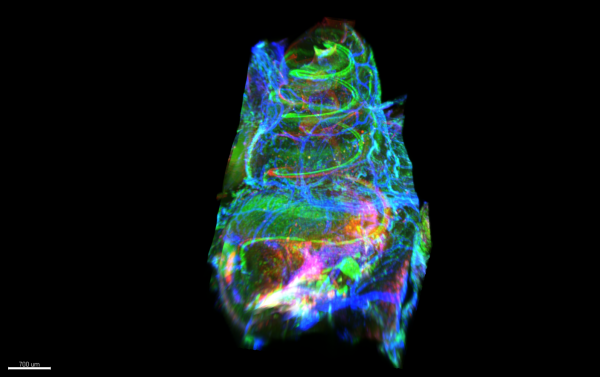
Cochlea in colour by Kate Brody, Ultramicroscope II
Title: Cochlea in colour by Kate Brody, Ultramicroscope II Description: Light sheet microscopy image showing a cochlea which is implanted with a cochlear implant. Hair cells and macrophages are labelled in the green, neurons and extracellular matrix in the red and arterioles and myo-fibroblasts in the blue. -
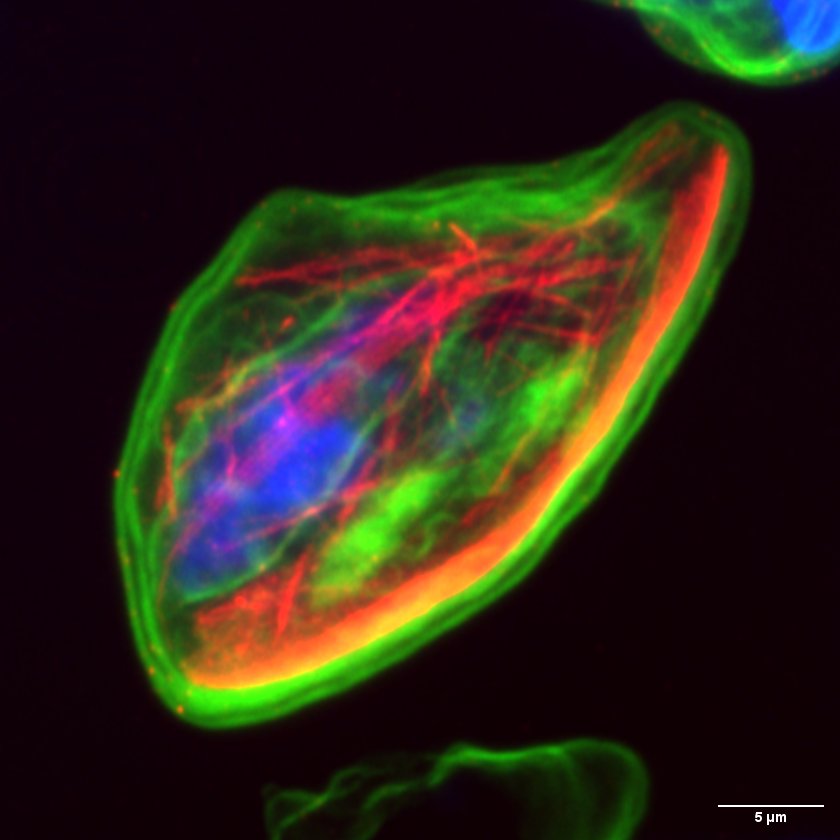
Plasmodium falciparum gametocyte microtubules by Jiahong Li, Elyra/LSM880
Title: Plasmodium falciparum gametocyte microtubules by Jiahong Li, Elyra/LSM880 Description: The unusual nuclear microtubules appear in the non-mitotic stage Plasmodium falciparum gametocyte. Understand the structure and function of nuclear microtubules can help us to block gametocyte development and malaria diseases transmission. The microtubules were labeled with anti-beta tubulin (red), the chromatins were labeled with DAPI, and the general proteins were stained with NHS-ester 488 dye. -
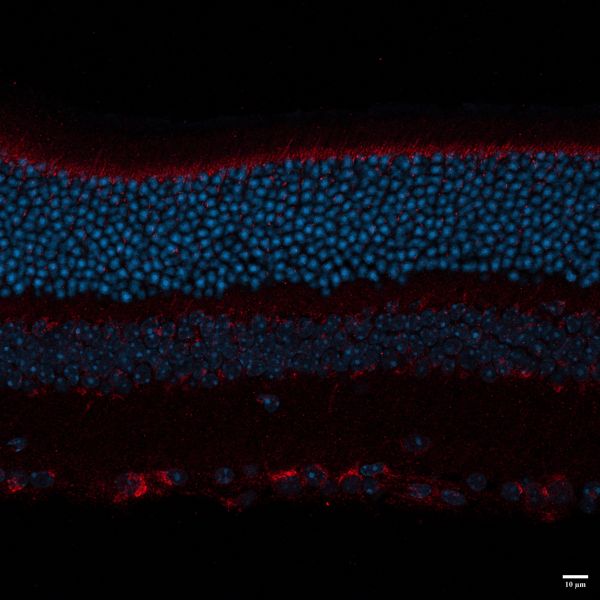
BLACK MOUSE P35 by Nadia Hosseini Naveh, LSM800
Title: BLACK MOUSE P35 by Nadia Hosseini Naveh, LSM800 Description: this image shows different layers of retina , I used LAMP1 for primary antibody , and 647 and DAPI for secondary . -
I 'Heart' Microscopy by Nicole Thomson, Nikon C2
Title: I 'Heart' Microscopy by Nicole Thomson, Nikon C2 Description: Arabidopsis thaliala heart stage embryo with gLUH nuclei expression -
Synaptic puncta on Hair Cells in the cochlea by Alexandra Hill, Nikon A1R
Title: Synaptic puncta on Hair Cells in the cochlea by Alexandra Hill, Nikon A1R Description: Here is a 60x coloc image of the IHC of a Cat cochlea, this was taken from a frequency point of 12kHz on a normal hearing cochlea. you can see the inner Hair cells i quiet vibrant and show Lots of synaptic Puncta on each cell. -
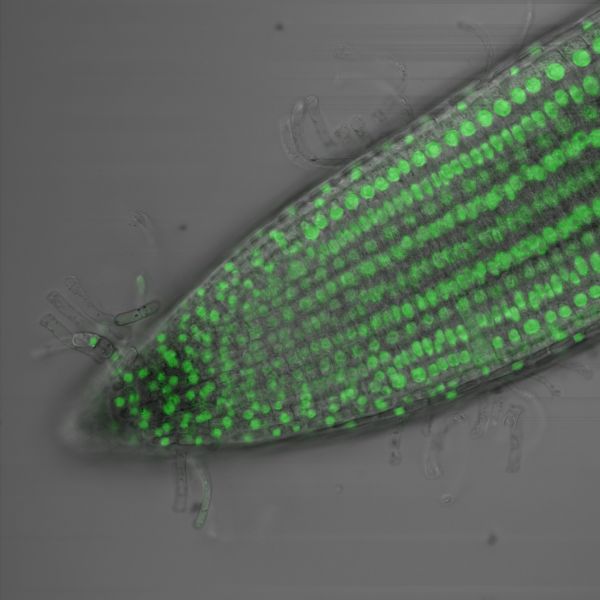
Green fluorescence in the root tip of Arabidopsis thaliana by Amelia Keynton, Nikon C2
Title: Green fluorescence in the root tip of Arabidopsis thaliana by Amelia Keynton, Nikon C2 Description: The green dots which represent the nucleus of the cell tell us that our fluorescent protein is being expressed in every cell in the root tip of Arabidopsis thaliana. We use this to determine where specifically in the root different genetic elements called promoters can drive protein expression. -
Peacock ganglia by Sapna Devi, Zeiss LSM780
Title: Peacock ganglia by Sapna Devi, Zeiss LSM780 Description: A collection of neurons and blood vessels key to functioning of the nervous system -
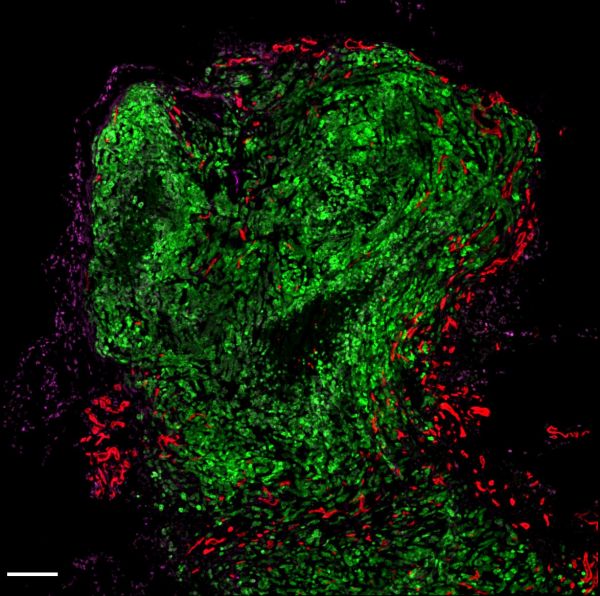
Green Alien by Sapna Devi, Zeiss LSM780
Title: Green Alien by Sapna Devi, Zeiss LSM780 Description: Tumour cells in green invading breast tissues surrounded by formation of new blood cells in red. Image shows how tumours expand and take over a normal tissue quickly. -
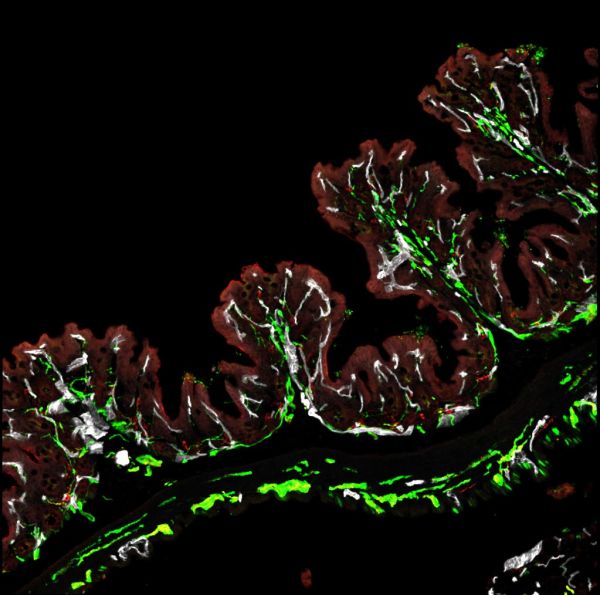
Ferny gut by Sapna Devi , Zeiss LSM780
Title: Ferny gut by Sapna Devi , Zeiss LSM780 Description: A typical structure found in the colon with lots of nerve innervation in green colocalizing with blood vessels in white. Image demonstrates how the body "talks" to the gut. -
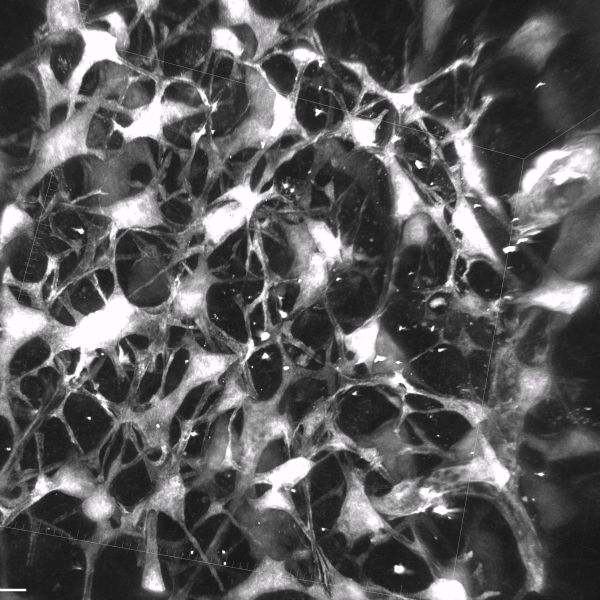
Traveling thru space by Sapna Devi, Zeiss LSM780
Title: Traveling thru space by Sapna Devi, Zeiss LSM780 Description: Cells known as fibroblastic reticular cells key in tissue architecture. -
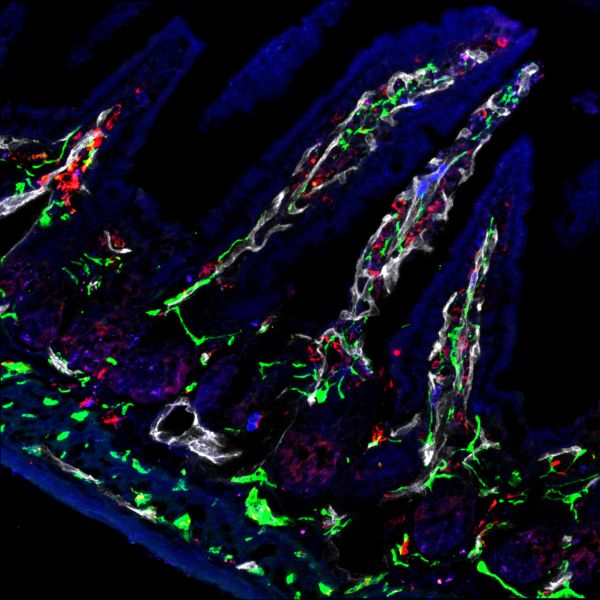
Gut feelings by Sapna Devi, Zeiss LSM780
Title: Gut feelings by Sapna Devi, Zeiss LSM780 Description: Nerves associated with blood vessels seen in the villus of the small intestine crucial for controlling gut functions. -
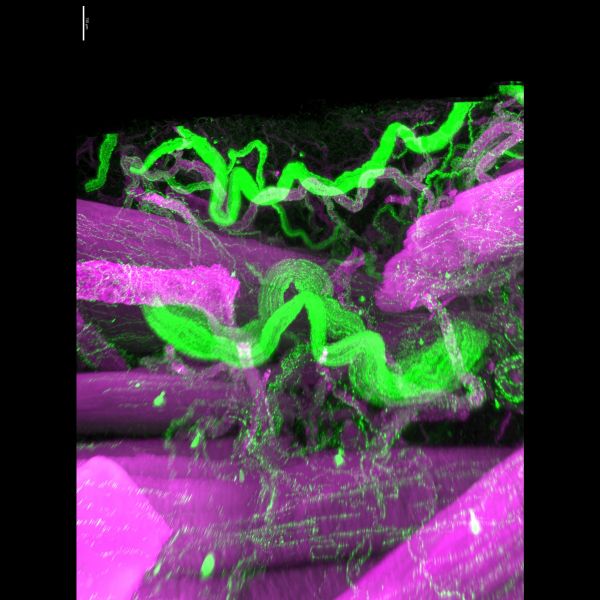
Nervous Break-in by Jonathan Chan, UMII
Title: Nervous Break-in by Jonathan Chan, UMII Description: A big axon (green) weaves in between a break in the smooth muscle layer (purple) in bladder serosa. A gentle reminder to take a well-deserved break when even when you have a wall of activities. -
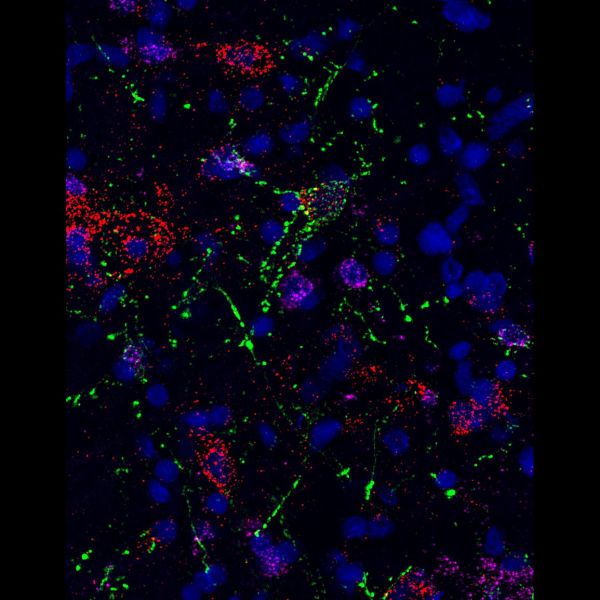
dot-to-dot by Angela Connelly, LSM880
Title: dot-to-dot by Angela Connelly, LSM880 Description: RNAscope image of the preBotzinger complex in the rat brainstem, with VGlut (Cy5), VGAT (Cy3) and GtACR2 opsin photoinhibited neurons in green. The neurochemistry of preBotzinger neurons which enable and coordinate breathing; which is vital for life, and have been inhibited via optogenetics, is great to understand. -
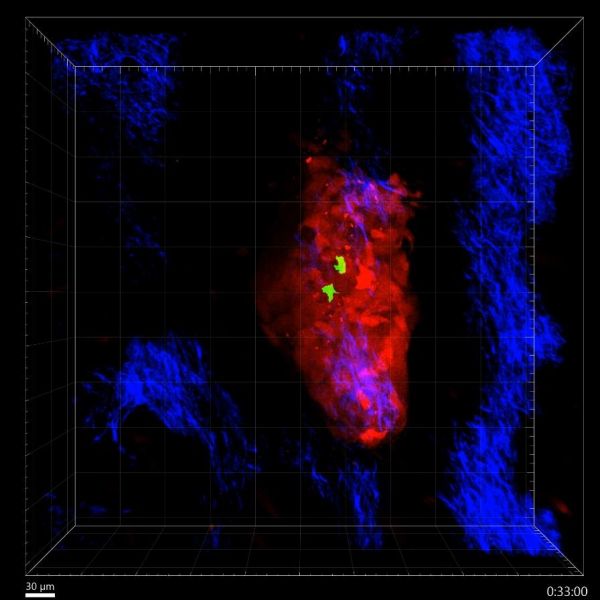
Well, well, well what do we have here? by Teagan Wagner, Olympus FVMPERS 2-Photon
Title: Well, well, well what do we have here?' T cells survey melanoma in the skin by Teagan Wagner, Olympus FVMPERS 2-Photon Microscope Description: Specialist immune cells, known as T cells, are important for recognising and fighting cancer. This video shows T cells (green) actively recognising and surveying a melanoma tumour (red) in the skin (blue) in 3-D in real-time for more than 2 hours. Video: https://youtu.be/zXcrkh908oI
Note: Image size was reduced for some images.